Our goal is to identify the brain regions most relevant to mental illness using neuroimaging. State of the art machine learning methods commonly suffer from repeatability difficulties in this application, particularly when using large and heterogeneous populations for samples. We revisit both dimensionality reduction and sparse modeling, and recast them in a common optimization-based framework. This allows us to combine the benefits of both types of methods in an approach which we call unambiguous components. We use this to estimate the image component with a constrained variability, which is best correlated with the unknown disease mechanism.
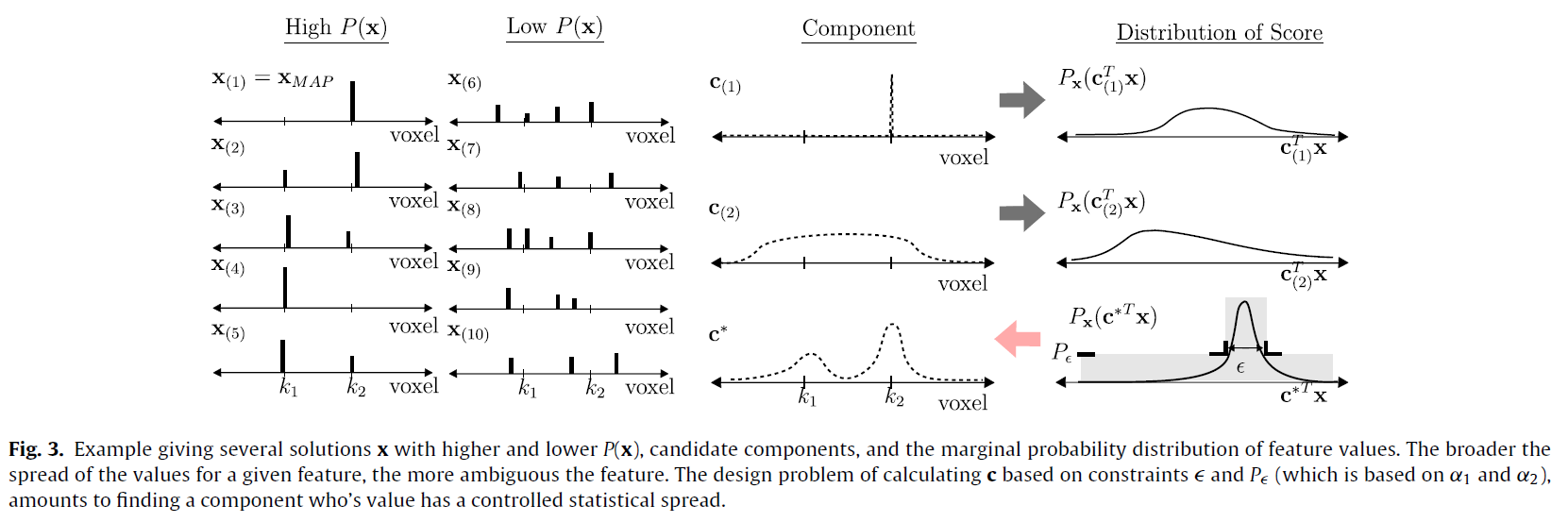
We apply the method to the estimation of neuroimaging biomarkers for schizophrenia, using task fMRI data from a large multi-site study. The proposed approach yields an improvement in both robustness of the estimate and classification accuracy. We find that unambiguous components incorporate roughly two thirds of the same brain regions as sparsity-based methods LASSO and elastic net, while roughly one third of the selected regions differ. Further, unambiguous components achieve superior classification accuracy in differentiating cases from controls.
K. Dillon, V. Calhoun, and Y.-P. Wang, “A robust sparse-modeling framework for estimating schizophrenia biomarkers from fMRI,”Journal of Neuroscience Methods, vol. 276, pp. 46–55, Jan. 2017. (pdf)