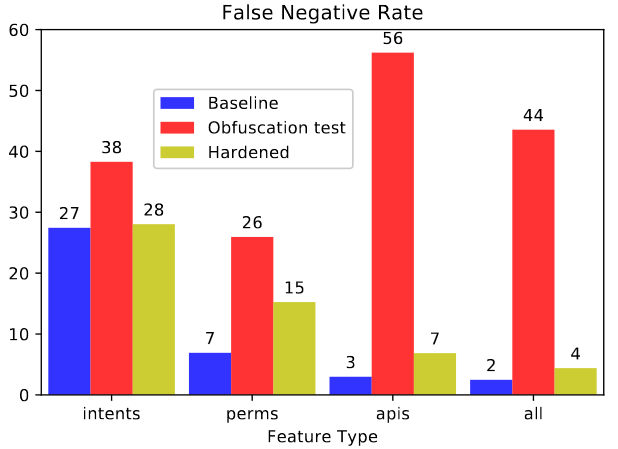
We consider the problem of detecting malware with deep learning models, where the malware may be combined with significant amounts of benign code. Examples of this include piggybacking and trojan horse attacks on a system, where malicious behavior is hidden within a useful application. Such added flexibility in augmenting the malware enables significantly more code obfuscation. Hence we focus on the use of static features, particularly Intents, Permissions, and API calls, which we presume cannot be ultimately hidden from the Android system, but only augmented with yet more such features. We first train a deep neural network classifier for malware classification using features of benign and malware samples. Then we demonstrate a steep increase in false negative rate (ie, attacks succeed), simply by randomly adding features of a benign app to malware. Finally we test the use of data augmentation to harden the classifier against such attacks. We find that for API calls, it is possible to reject the vast majority of attacks, where using Intents or Permissions is less successful.